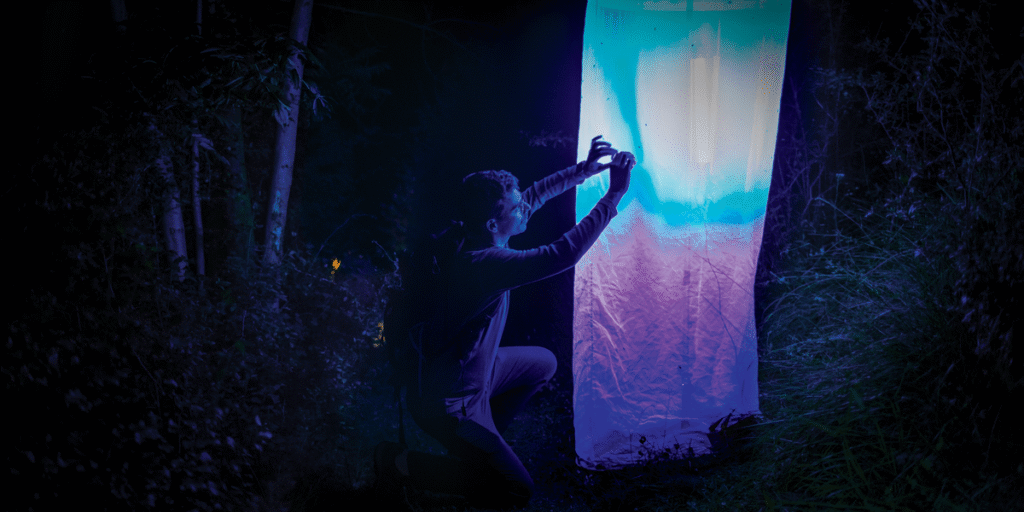A gibbous moon hangs over a lonely mountain path within the Italian Alps, above the village of Malles Venosta, whose lights dot the valley beneath. Benjamin Wiesmair stands subsequent to a moth entice as tall as he’s, his face, bushy beard, and hair bun lit by its purple glow. He’s carrying a headlamp, a dusty and battered smartwatch, cargo shorts, and a blue zip sweater with the sleeves pulled up. Numerous moths beat frenetically across the entice’s white, diaphanous panels, that are swaying with ghostly ripples in a mild breeze. Wiesmair squints at his smartphone, which is logged on to a database of European moth species.
“Chersotis multangula,” he says.
“Sure, we want that,” comes the crisp reply from Clara Spilker, consulting a laptop computer.
Wiesmair, an entomologist on the Tyrolean State Museums, in Innsbruck, Austria, and Spilker, a technical assistant on the Senckenberg German Entomological Institute, in Müncheberg, are collaborating in some of the far-reaching organic initiatives ever: acquiring a genome sequence for practically each named species of eukaryotic organism on the planet. All 1.8 million of them. The researchers are a part of an expedition for Project Psyche, which is sampling European butterflies and moths and can feed its information into the worldwide initiative, known as the Earth BioGenome Project (EBP).
Entomologist Benjamin Wiesmair [at right] makes use of his smartphone to seek the advice of a lepidoptera database to establish the species of moths captured throughout a trapping session on an alpine path above Malles Venosta, Italy. Clara Spilker and Alena Sucháčková [middle] seek the advice of a desk to find out whether or not the species are wanted for genome sequencing.
Luigi Avantaggiato
Eukaryotes are organisms whose cells include a nucleus. From protozoa to human beings, all have the identical primary organic mechanism for constructing, sustaining, and propagating their type of life: a genome. It’s the sum whole of the genes carried by the creature.
Twenty-two years in the past, researchers introduced that for the primary time they’d mapped, or “sequenced,” practically the entire genes in a human genome. The venture price more than US $3 billion and took 13 years, however it will definitely reworked medical follow. Within the new period of genomic medicine, medical doctors can take a affected person’s particular genetic make-up into consideration throughout prognosis and therapy.

Many moths, interested in the ultraviolet lights, had been captured throughout a light-trapping tour close to Malles Venosta, Italy.
Luigi Avantaggiato
The EBP goals to succeed in its monumental objective by 2035. As of July 2024, its tally of genomes sequenced stood at about 4,200. Success will undoubtedly rely on researchers’ capability to scale a number of biotech applied sciences.
“We have to scale, from the place we’re at, greater than a hundredfold by way of the variety of genomes per yr that we’re producing worldwide,” says Harris Lewin, who leads the EBP and is a professor and genetics researcher at Arizona State University.
Probably the most essential applied sciences that should be scaled is a way known as long-read genome sequencing. Specialists on the entrance traces of the genomic revolution in biology are assured that such scaling might be doable, their conviction coming partly from previous expertise. “In comparison with 2001,” when the Human Genome Project was nearing completion, “it’s now roughly 500,000 occasions cheaper to sequence DNA,” says Steven Salzberg, a Bloomberg Distinguished Professor at Johns Hopkins University and director of the varsity’s Center for Computational Biology. “And additionally it is about 500,000 occasions sooner to sequence,” he provides. “That’s the scale, over the previous 25 years, a scale of acceleration that has vastly outstripped any enhancements in computational expertise, both in reminiscence or velocity of processors.”

A lepidopterist wrote figuring out data on a label affixed to a specimen jar containing a moth captured throughout a light-trapping tour close to Malles Venosta, Italy.
Luigi Avantaggiato
There are a lot of causes to cheer on the EBP and the technological advances that can underpin it. Having established a genome for each eukaryotic creature, researchers will acquire deep new insights into the connections among the many threads in Earth’s internet of life, and into how evolution proceeded for its myriad life kinds. That data will turn into more and more essential as climate change alters the ecosystems on which all of these creatures, together with us, rely.
And though the venture is a scientific collaboration, it might spin off sizable monetary windfalls. Many medication, enzymes, catalysts, and different chemical substances of incalculable worth had been first identified in natural samples. Researchers anticipate many extra to be found within the means of figuring out, in impact, every of the billions of eukaryotic genes on Earth, a lot of which encode a protein of some sort.
“One thought is that by taking a look at crops, which have all types of chemical substances, typically which they make so as to combat off insects or pests, we would discover new molecules which might be going to be essential medication,” says Richard Durbin, professor of genetics on the University of Cambridge and a veteran of a number of genome sequencing initiatives. The immunosuppressant and cancer drug rapamycin, to quote simply one among numerous examples, got here from a microbe genome.
Your Genes Are a Large Cause Why You’re You
The EBP is an umbrella group for some 60 projects (and counting) which might be sequencing species in both a area or in a specific taxonomic group. The overachiever is the Darwin Tree of Life Project, which is sequencing all species in Britain and Ireland, and has contributed about half of the entire genomes recorded by the EBP up to now. Mission Psyche was spun out of the Darwin Tree of Life initiative, and each have obtained beneficiant help from the Wellcome Trust.
To get an thought of the magnitude of the general EBP, think about what it takes to sequence a species. First, an organism should be discovered or captured and sampled, after all. That’s what introduced Wiesmair, Spilker, and 41 different lepidopterists to the Italian Alps for the Mission Psyche expedition this previous July. Over 5 days, they collected greater than 200 new species for sequencing, which can increase the 1,000 completed lepidoptera genome sequences already accomplished and the roughly 2,000 samples awaiting sequencing. There’s nonetheless loads of work to be achieved; there are round 11,000 species of moths and butterflies throughout Europe and Britain.
After sampling, genetic materials—the creature’s DNA—is collected from cells after which damaged up into fragments which might be brief sufficient to be learn by the sequencing machines. After sequencing, the genome information is analyzed to find out the place the genes are and, if doable, what they do.
Over the previous 25 years, the acceleration of gene-sequencing tech has vastly outstripped any enhancements in computational expertise, both in reminiscence or velocity of processors.
DNA is a molecule whose construction is the well-known double helix. It resides within the nucleus of each cell within the physique of each dwelling factor. For those who consider the molecule as a twisted ladder, the rungs of the ladder are shaped by pairs of chemical models known as bases. There are 4 completely different bases: adenine (A), guanine (G), cytosine (C), and thymine (T). Adenine all the time pairs with thymine, and guanine all the time pairs with cytosine. So a “rung” may be any of 4 issues: A–T, T–A, C–G, or G–C.
These 4 base-pair permutations are the symbols that comprise the code of life. Strings of them make up the genome as segments of varied lengths known as genes. Your genes at the very least partially management most of your bodily and plenty of of your psychological traits—not solely what coloration your eyes are and the way tall you’re but in addition what illnesses you’re vulnerable to, how tough it’s so that you can construct muscle or shed weight, and even whether or not you’re liable to motion sickness.
How Lengthy-Learn Genome Sequencing Works
Lengthy-read sequencing begins by breaking apart a pattern of genetic materials into items which might be typically about 20,000 base pairs lengthy. Then the sequencing expertise reads the sequence of base pairs on these DNA strands to provide random segments, known as “reads,” of DNA which might be at the very least 10,000 pairs in size. As soon as these lengthy reads are obtained, highly effective bioinformatics software program is used to build longer stretches of contiguous sequence by overlapping reads that share the identical sequence of bases.
To grasp the method, consider a genome as a novel, and every of its separate chromosomes as a chapter within the novel. Think about shredding the novel into items of paper, every about 5 sq. centimeters. Your job is to reassemble them into the unique novel (sadly for you, the pages aren’t numbered). What makes this process doable is overlap—you shredded a number of copies of the novel, and the items overlap, making it simpler to see the place one leaves off and one other begins.
Making it a lot tougher, nonetheless, are the various sections of the ebook stuffed with repetitive nonsense: the identical phrase repeated a whole lot and even hundreds of occasions. A minimum of half of a typical mammalian genome consists of those repetitive sequences, a few of which have regulatory functions and others thought to be “junk” DNA that’s descended from historical genes or viral infections and not useful. Lengthy-read expertise is adept at dealing with these repetitive sequences. Going again to the novel-shredding analogy, think about attempting to reassemble the ebook after it was shredded into items only one centimeter sq. relatively than 5. That’s analogous to the problem that researchers previously confronted attempting to assemble million-base-pair DNA sequences utilizing older, “short-read” sequencing technology.
The Two Approaches to Lengthy-Learn Sequencing
The long-read sequencing market has two main corporations—Oxford Nanopore Technologies (ONT) and Pacific Biosciences of California (PacBio)—which compete intensely. The 2 corporations have developed totally completely different programs.
The center of ONT’s system is a circulate cell that accommodates 2,000 or extra extraordinarily tiny apertures known as, appropriately sufficient, nanopores. The nanopores are anchored in an electrically resistant membrane, which is built-in onto a sensor chip. In operation, every finish of a phase of DNA is hooked up to a molecule known as an adapter that accommodates a helicase enzyme. A voltage is utilized throughout the nanopore to create an electric field, and the sector captures the DNA with the hooked up adapter. The helicase begins to unzip the double-stranded DNA, with one of many DNA strands passing via the nanopore, base by base, and the opposite launched into the medium.
What propels the strand via the nanopore is that voltage—it’s solely about 0.2 volts, however the nanopore is barely 5 nanometers broad, so the electrical area is a number of hundred thousand volts per meter. “It’s like a flash of lightning going via the pore,” says David Deamer, one of many inventors of the expertise. “At first, we had been afraid we might fry the DNA, nevertheless it turned out that the encircling water absorbed the warmth.”
That sort of area energy would ordinarily propel the DNA-based molecule via the pore at speeds far too quick for evaluation. However the helicase acts like a brake, inflicting the molecule to undergo with a ratcheting movement, one base at a time, at a still-lively fee of about 400 bases per second. In the meantime, the electrical area additionally propels a circulate of ions throughout the nanopore. This present circulate is decreased by the presence of a base within the nanopore—and, crucially, the quantity of the lower is determined by which of the 4 bases, A, T, G, or C, is getting into the pore. The result’s {an electrical} sign that may be quickly translated right into a sequence of bases.
PacBio’s machines depend on an optical relatively than an digital technique of figuring out the bases. PacBio’s latest process, which it calls HiFi, begins by capping each ends of the DNA phase and untwisting it to create a single-stranded loop. Every loop is then positioned in an infinitesimally tiny nicely in a microchip, which might have 25 million of these wells. Hooked up to every loop is a polymerase enzyme, which serves a crucial perform each time a cell divides. It attaches to single-stranded DNA and provides the complementary bases, making every rung of the ladder entire once more. PacBio makes use of particular variations of the 4 bases which were engineered to fluoresce in a attribute coloration when uncovered to ultraviolet light.
A UV laser shines via the underside of the tiny nicely, and a photosensor on the high detects the faint flashes of sunshine because the polymerase goes across the DNA pattern loop, base by base. The upshot is that there’s a sequence of sunshine flashes, at a fee of about three per second, that reveals the sequence of base pairs within the DNA pattern.
As a result of the DNA pattern has been transformed right into a loop, the entire course of may be repeated, to realize increased accuracy, by merely going across the loop one other time. PacBio’s flagship Revio machine usually makes 5 to 10 passes, reaching median accuracy charges as excessive as 99.9 %, based on Aaron Wenger, senior director of product advertising and marketing on the firm.
How Researchers Will Scale Up Lengthy-Learn Sequencing
That sort of accuracy doesn’t come low cost. A Revio system, which has 4 chips, every with 25 million wells, prices round $600,000, based on Wenger. It weighs 465 kilograms and is concerning the measurement of a giant household fridge. PacBio says a single Revio can sequence about 4 total human genomes in a 24-hour interval for lower than $1,000 per genome.
ONT claims accuracy above 99 % for its flagship machine, known as PromethION 24. It prices round $300,000, based on Rosemary Sinclair Dokos, chief product and advertising and marketing officer at ONT. One other benefit of the ONT PromethION system is its capability to course of fragments of DNA with as many as one million base pairs. ONT additionally provides an entry-level system, known as MinION Mk1D, for simply $3,000. It’s concerning the measurement of two smartphones stacked on high of one another, and it plugs right into a laptop computer, providing researchers a setup that may simply be toted into the sector.

On the Centro Nacional de Análisis Genómico, in Barcelona, technician Álvaro Carreras prepares a PromethION long-read sequencing machine, from Oxford Nanopore Applied sciences, to sequence a genome. Behind Carreras is a Pacific Biosciences Revio long-read machine.
Luigi Avantaggiato
Though researchers typically have sturdy preferences, it’s not unusual for a state-of-the-art genetics laboratory to be geared up with machines from each corporations. At Barcelona’s Centro Nacional de Análisis Genómico, for instance, researchers have entry to each PacBio Revio machines in addition to PromethION 24 and GridION machines from ONT.
Durbin, at Cambridge University, sees numerous upside within the present state of affairs. “It’s superb to have two corporations,” he declares. “They’re in competitors with one another for the market.” And that competitors will undoubtedly gas the tech advances that the EBP’s backers are relying on to get the venture throughout the end line.

A technician on the Centro Nacional de Análisis Genómico, in Barcelona, holds a circulate cell for a PromethION long-read sequencing machine from Oxford Nanopore Applied sciences. The circulate cell accommodates a chip that interacts with the pattern of DNA to carry out the long-read sequencing.
Luigi Avantaggiato
PacBio’s Wenger notes that the 25-million-well chips that underpin its Revio system are nonetheless being fabricated on 200-millimeter semiconductor wafers. A transfer to 300-mm wafers and extra superior lithographic methods, he says, would allow them to get many extra chips per wafer and put a whole lot of thousands and thousands of wells on every of these chips—if the market calls for it.
At ONT, Dokos describes comparable math. A single circulate cell now consists of greater than 2,000 nanopores, and a state-of-the-art PromethION 24 system can have 24 circulate cells (or upward of 48,000 nanopores) operating in parallel. However a future system might have a whole lot of hundreds of nanopores, she says—once more, if the market calls for it.
The EBP will want all of these advances, and extra. EBP director Lewin notes that after seven years, the three-phase initiative is wrapping up part one and getting ready for part two. The objective for part two is to sequence 150,000 genomes between 2026 and 2030. For part two, “We’ve received to get to 37,500 genomes per yr,” Lewin says. “Proper now, we’re getting shut to three,000 per yr.” In part two, the price per genome sequenced may also have to say no from roughly $26,000 per genome in part one to $6,100, based on the EBP’s official road map. That $6,100 determine consists of all prices—not simply sequencing but in addition sampling and the opposite levels wanted to provide a completed genome, with the entire genes recognized and assigned to chromosomes.

A technician on the Centro Nacional de Análisis Genómico, in Barcelona, introduces a pattern of fragmented DNA for sequencing in a PromethION machine from Oxford Nanopore Applied sciences.
Luigi Avantaggiato
Part three will up the ante even increased. The highway map requires greater than 1.65 million genome sequences between 2030 and 2035 at a price of $1,900 per genome. If they will pull it off, the complete venture may have price roughly $4.7 billion—significantly much less in actual phrases than what it price to do exactly the human genome 22 years in the past. The entire information collected—the genome sequences for all named species on Earth—will occupy a bit over 1 exabyte (1 billion gigabytes) of digital storage.
It is going to arguably be probably the most worthwhile exabyte in all of science. “With this genomic information, we will get to one of many questions that Darwin requested a very long time in the past, which is, How does a species come up? What’s the origin of species? That’s his well-known ebook the place he by no means really answered the query,” says Mark Blaxter, who leads the Darwin Tree of Life Mission on the Wellcome Sanger Institute close to Cambridge and who additionally conceived and began Mission Psyche. “We’ll get a a lot, a lot better thought about what it’s that makes a species and the way species are distinct from one another.”
A portion of that data will come from the various moths collected on these summer season nights within the Italian Alps. Lepidoptera “return round 300 million years,” says Charlotte Wright, a co-leader, together with Blaxter, of Mission Psyche. Analyzing the genomes of big numbers of species will assist clarify why some branches of the lepidoptera have advanced much more species than others, she says.
And that sort of data ought to finally accumulate into solutions to a few of biology’s most profound questions on evolution and the mechanisms by which it acts. “The wonderful factor is that by doing this for the entire lepidoptera of Europe, we aren’t simply studying about particular person circumstances,” says Wright. “We’ve realized throughout all of it.”
From Your Website Articles
Associated Articles Across the Net